Note
Click here to download the full example code
Numpy: arrays and matrices¶
NumPy is an extension to the Python programming language, adding support for large, multi-dimensional (numerical) arrays and matrices, along with a large library of high-level mathematical functions to operate on these arrays.
Sources:
Kevin Markham: https://github.com/justmarkham
Computation time:
import numpy as np
l = [v for v in range(10 ** 8)] s = 0 %time for v in l: s += v
arr = np.arange(10 ** 8) %time arr.sum()
Create arrays¶
Create ndarrays from lists. note: every element must be the same type (will be converted if possible)
import numpy as np
data1 = [1, 2, 3, 4, 5] # list
arr1 = np.array(data1) # 1d array
data2 = [range(1, 5), range(5, 9)] # list of lists
arr2 = np.array(data2) # 2d array
arr2.tolist() # convert array back to list
Out:
[[1, 2, 3, 4], [5, 6, 7, 8]]
create special arrays
np.zeros(10)
np.zeros((3, 6))
np.ones(10)
np.linspace(0, 1, 5) # 0 to 1 (inclusive) with 5 points
np.logspace(0, 3, 4) # 10^0 to 10^3 (inclusive) with 4 points
Out:
array([ 1., 10., 100., 1000.])
arange is like range, except it returns an array (not a list)
int_array = np.arange(5)
float_array = int_array.astype(float)
Examining arrays¶
arr1.dtype # float64
arr2.ndim # 2
arr2.shape # (2, 4) - axis 0 is rows, axis 1 is columns
arr2.size # 8 - total number of elements
len(arr2) # 2 - size of first dimension (aka axis)
Out:
2
Reshaping¶
arr = np.arange(10, dtype=float).reshape((2, 5))
print(arr.shape)
print(arr.reshape(5, 2))
Out:
(2, 5)
[[0. 1.]
[2. 3.]
[4. 5.]
[6. 7.]
[8. 9.]]
Add an axis
a = np.array([0, 1])
a_col = a[:, np.newaxis]
print(a_col)
#or
a_col = a[:, None]
Out:
[[0]
[1]]
Transpose
print(a_col.T)
Out:
[[0 1]]
Flatten: always returns a flat copy of the orriginal array
arr_flt = arr.flatten()
arr_flt[0] = 33
print(arr_flt)
print(arr)
Out:
[33. 1. 2. 3. 4. 5. 6. 7. 8. 9.]
[[0. 1. 2. 3. 4.]
[5. 6. 7. 8. 9.]]
Ravel: returns a view of the original array whenever possible.
arr_flt = arr.ravel()
arr_flt[0] = 33
print(arr_flt)
print(arr)
Out:
[33. 1. 2. 3. 4. 5. 6. 7. 8. 9.]
[[33. 1. 2. 3. 4.]
[ 5. 6. 7. 8. 9.]]
Summary on axis, reshaping/flattening and selection¶
Numpy internals: By default Numpy use C convention, ie, Row-major language: The matrix is stored by rows. In C, the last index changes most rapidly as one moves through the array as stored in memory.
For 2D arrays, sequential move in the memory will:
- iterate over rows (axis 0)
iterate over columns (axis 1)
For 3D arrays, sequential move in the memory will:
- iterate over plans (axis 0)
- iterate over rows (axis 1)
iterate over columns (axis 2)
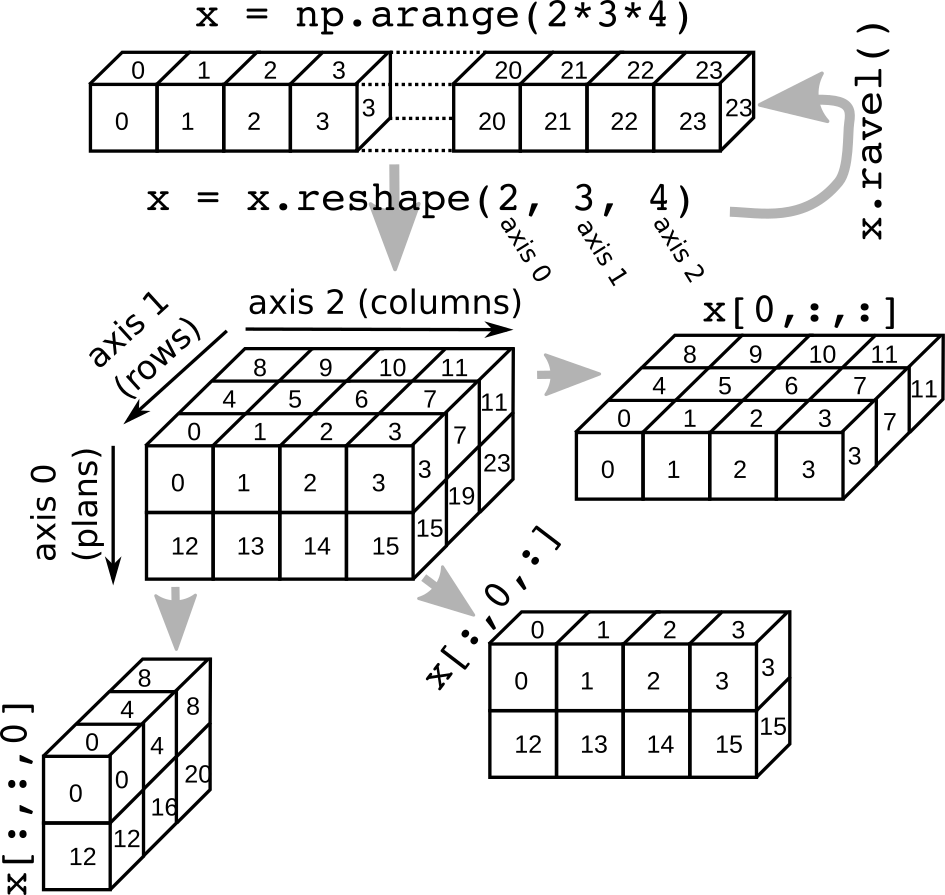
x = np.arange(2 * 3 * 4)
print(x)
Out:
[ 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23]
Reshape into 3D (axis 0, axis 1, axis 2)
x = x.reshape(2, 3, 4)
print(x)
Out:
[[[ 0 1 2 3]
[ 4 5 6 7]
[ 8 9 10 11]]
[[12 13 14 15]
[16 17 18 19]
[20 21 22 23]]]
Selection get first plan
print(x[0, :, :])
Out:
[[ 0 1 2 3]
[ 4 5 6 7]
[ 8 9 10 11]]
Selection get first rows
print(x[:, 0, :])
Out:
[[ 0 1 2 3]
[12 13 14 15]]
Selection get first columns
print(x[:, :, 0])
Out:
[[ 0 4 8]
[12 16 20]]
Simple example with 2 array
Exercise:
Get second line
Get third column
arr = np.arange(10, dtype=float).reshape((2, 5))
print(arr)
arr[1, :]
arr[:, 2]
Out:
[[0. 1. 2. 3. 4.]
[5. 6. 7. 8. 9.]]
array([2., 7.])
Ravel
print(x.ravel())
Out:
[ 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23]
Stack arrays¶
a = np.array([0, 1])
b = np.array([2, 3])
Horizontal stacking
np.hstack([a, b])
Out:
array([0, 1, 2, 3])
Vertical stacking
np.vstack([a, b])
Out:
array([[0, 1],
[2, 3]])
Default Vertical
np.stack([a, b])
Out:
array([[0, 1],
[2, 3]])
Selection¶
Single item
arr = np.arange(10, dtype=float).reshape((2, 5))
arr[0] # 0th element (slices like a list)
arr[0, 3] # row 0, column 3: returns 4
arr[0][3] # alternative syntax
Out:
3.0
Slicing¶
Syntax: start:stop:step with start (default 0) stop (default last) step (default 1)
arr[0, :] # row 0: returns 1d array ([1, 2, 3, 4])
arr[:, 0] # column 0: returns 1d array ([1, 5])
arr[:, :2] # columns strictly before index 2 (2 first columns)
arr[:, 2:] # columns after index 2 included
arr2 = arr[:, 1:4] # columns between index 1 (included) and 4 (excluded)
print(arr2)
Out:
[[1. 2. 3.]
[6. 7. 8.]]
Slicing returns a view (not a copy) Modification
arr2[0, 0] = 33
print(arr2)
print(arr)
Out:
[[33. 2. 3.]
[ 6. 7. 8.]]
[[ 0. 33. 2. 3. 4.]
[ 5. 6. 7. 8. 9.]]
Row 0: reverse order
print(arr[0, ::-1])
# The rule of thumb here can be: in the context of lvalue indexing (i.e. the indices are placed in the left hand side value of an assignment), no view or copy of the array is created (because there is no need to). However, with regular values, the above rules for creating views does apply.
Out:
[ 4. 3. 2. 33. 0.]
Fancy indexing: Integer or boolean array indexing¶
Fancy indexing returns a copy not a view.
Integer array indexing
arr2 = arr[:, [1, 2, 3]] # return a copy
print(arr2)
arr2[0, 0] = 44
print(arr2)
print(arr)
Out:
[[33. 2. 3.]
[ 6. 7. 8.]]
[[44. 2. 3.]
[ 6. 7. 8.]]
[[ 0. 33. 2. 3. 4.]
[ 5. 6. 7. 8. 9.]]
Boolean arrays indexing
arr2 = arr[arr > 5] # return a copy
print(arr2)
arr2[0] = 44
print(arr2)
print(arr)
Out:
[33. 6. 7. 8. 9.]
[44. 6. 7. 8. 9.]
[[ 0. 33. 2. 3. 4.]
[ 5. 6. 7. 8. 9.]]
However, In the context of lvalue indexing (left hand side value of an assignment) Fancy authorizes the modification of the original array
arr[arr > 5] = 0
print(arr)
Out:
[[0. 0. 2. 3. 4.]
[5. 0. 0. 0. 0.]]
Boolean arrays indexing continues
names = np.array(['Bob', 'Joe', 'Will', 'Bob'])
names == 'Bob' # returns a boolean array
names[names != 'Bob'] # logical selection
(names == 'Bob') | (names == 'Will') # keywords "and/or" don't work with boolean arrays
names[names != 'Bob'] = 'Joe' # assign based on a logical selection
np.unique(names) # set function
Out:
array(['Bob', 'Joe'], dtype='<U4')
Vectorized operations¶
nums = np.arange(5)
nums * 10 # multiply each element by 10
nums = np.sqrt(nums) # square root of each element
np.ceil(nums) # also floor, rint (round to nearest int)
np.isnan(nums) # checks for NaN
nums + np.arange(5) # add element-wise
np.maximum(nums, np.array([1, -2, 3, -4, 5])) # compare element-wise
# Compute Euclidean distance between 2 vectors
vec1 = np.random.randn(10)
vec2 = np.random.randn(10)
dist = np.sqrt(np.sum((vec1 - vec2) ** 2))
# math and stats
rnd = np.random.randn(4, 2) # random normals in 4x2 array
rnd.mean()
rnd.std()
rnd.argmin() # index of minimum element
rnd.sum()
rnd.sum(axis=0) # sum of columns
rnd.sum(axis=1) # sum of rows
# methods for boolean arrays
(rnd > 0).sum() # counts number of positive values
(rnd > 0).any() # checks if any value is True
(rnd > 0).all() # checks if all values are True
# random numbers
np.random.seed(12234) # Set the seed
np.random.rand(2, 3) # 2 x 3 matrix in [0, 1]
np.random.randn(10) # random normals (mean 0, sd 1)
np.random.randint(0, 2, 10) # 10 randomly picked 0 or 1
Out:
array([0, 0, 0, 1, 1, 0, 1, 1, 1, 1])
Broadcasting¶
Sources: https://docs.scipy.org/doc/numpy-1.13.0/user/basics.broadcasting.html Implicit conversion to allow operations on arrays of different sizes. - The smaller array is stretched or “broadcasted” across the larger array so that they have compatible shapes. - Fast vectorized operation in C instead of Python. - No needless copies.
Rules¶
Starting with the trailing axis and working backward, Numpy compares arrays dimensions.
If two dimensions are equal then continues
If one of the operand has dimension 1 stretches it to match the largest one
When one of the shapes runs out of dimensions (because it has less dimensions than the other shape), Numpy will use 1 in the comparison process until the other shape’s dimensions run out as well.
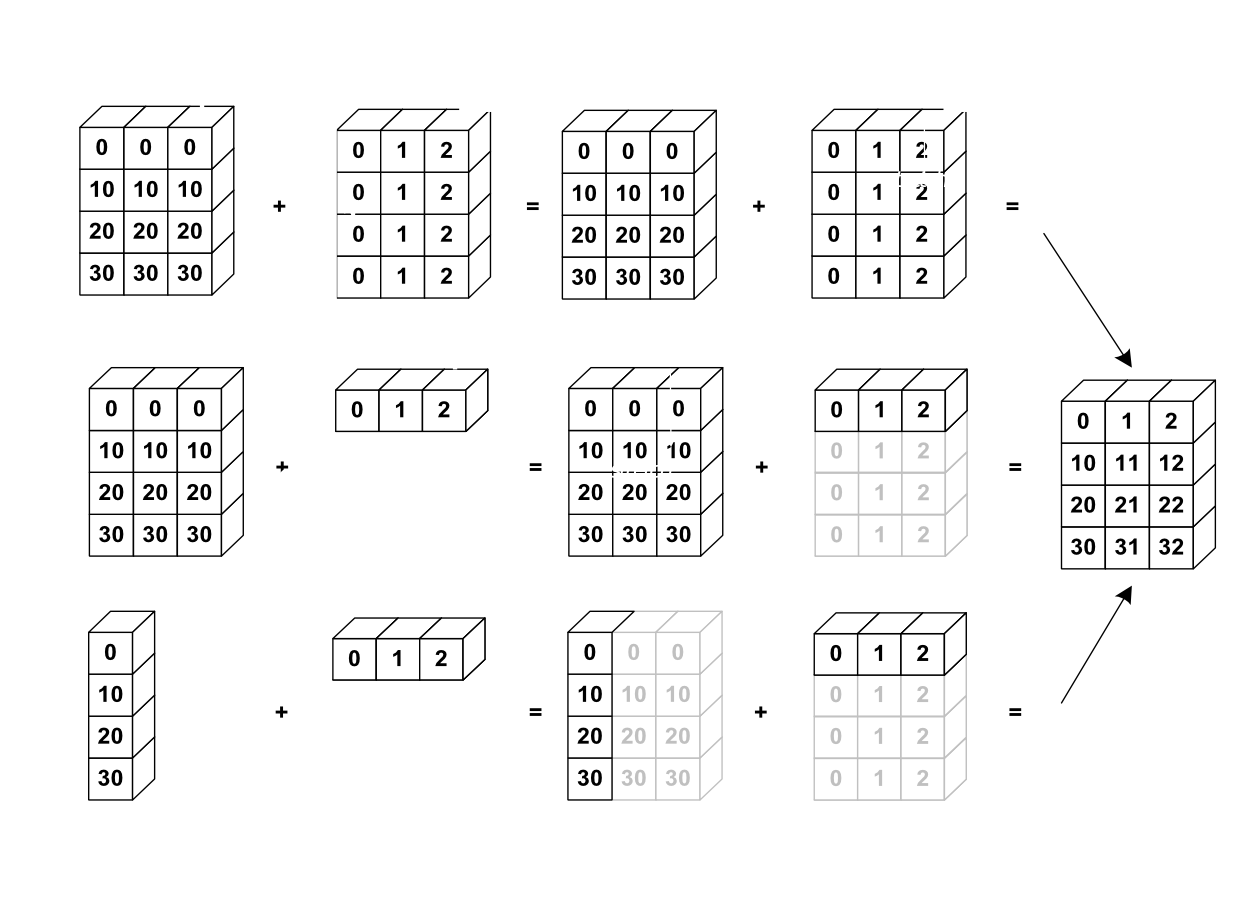
Source: http://www.scipy-lectures.org¶
a = np.array([[ 0, 0, 0],
[10, 10, 10],
[20, 20, 20],
[30, 30, 30]])
b = np.array([0, 1, 2])
print(a + b)
Out:
[[ 0 1 2]
[10 11 12]
[20 21 22]
[30 31 32]]
Center data column-wise
a - a.mean(axis=0)
Out:
array([[-15., -15., -15.],
[ -5., -5., -5.],
[ 5., 5., 5.],
[ 15., 15., 15.]])
Scale (center, normalise) data column-wise
(a - a.mean(axis=0)) / a.std(axis=0)
Out:
array([[-1.34164079, -1.34164079, -1.34164079],
[-0.4472136 , -0.4472136 , -0.4472136 ],
[ 0.4472136 , 0.4472136 , 0.4472136 ],
[ 1.34164079, 1.34164079, 1.34164079]])
Examples
Shapes of operands A, B and result:
A (2d array): 5 x 4
B (1d array): 1
Result (2d array): 5 x 4
A (2d array): 5 x 4
B (1d array): 4
Result (2d array): 5 x 4
A (3d array): 15 x 3 x 5
B (3d array): 15 x 1 x 5
Result (3d array): 15 x 3 x 5
A (3d array): 15 x 3 x 5
B (2d array): 3 x 5
Result (3d array): 15 x 3 x 5
A (3d array): 15 x 3 x 5
B (2d array): 3 x 1
Result (3d array): 15 x 3 x 5
Exercises¶
Given the array:
X = np.random.randn(4, 2) # random normals in 4x2 array
For each column find the row index of the minimum value.
Write a function
standardize(X)that return an array whose columns are centered and scaled (by std-dev).
Total running time of the script: ( 0 minutes 0.012 seconds)